前号以来今日まで即ち平成9年3月から平成9年7月までの活動報告を行う。 平成8年度後期のビームタイム終了直後である平成8年12月16日より放射 光実験施設は低エミッタンス化のため平成9年10月までの大規模な工事に入っ た。そのため今回利用実験の報告は無い。今期はTARA用可搬型コンテナハウ ス(TARA用プレハブ)の2階がほぼ完成したこと及び計算機やネットワーク、 将来計画等及び本プロジェクトの論文に関する記事等を報告する。
本年4月1日をもって「高エネルギー物理学研究所」が「高エネルギー加速器 研究機構」に組織替えされ、それに伴い「放射光実験施設」は「物質構造科学研 究所」になった。略称は「物構研」である。TARAとの関係は名称の変更以外 変わりなく、これまで通り継続する。KEK及びPFという略称は今後とも残さ れるが物構研には中性子線関連の部門も含まれるため放射光については「放射光 実験施設」ではなく「放射光施設」となった。
また本年3月81日を持って本プロジェクトの代表であった坂部知平が定年に より退官した。研究プロジェクトの代表は筑波大学の教官に限るというTARA の規則によりプロジェクト期間内ではあるが、坂部知平が代表を務めることはで きなくなった。後任には筑波大学応用生物化学系教授祥雲弘文氏が引き受けて下 さった。従って今後筑波大学内のこと及び物構研との正式書類は全て祥雲弘文教 授の名前で行われる。今後メンパーに直接関係することとしては実験の際提出す る派遣届けの責任者の欄は祥雲弘文(TARA)となる。坂部プロジェクトとい う略称は今後とも残され、上記以外のことは今後とも坂部知平が行う。
I. TARA用実験ステーション
1. BL6B
TARA用実験ステーションBL6Bの大きな変更はない。
利用者用マニュアルは本号58頁を参照。
2. 大型IP読取装置
前回のビームタイム中発生した大型IP読取装置のトラブルは理学電機が対処 し可成り減少した。中丸幸雄氏の報告によると、念入りな調整の結果IP搬送系 のエラーは1,000回のテストに対し全く異常は見付からない状態になった。 又、読取中lNDYの動作が極めて遅くなり使用不能になる現象は電源電圧を上げる などの処置により極めて減少した。1,000回のテスト中全く正常な装置もあ る。スカジーエラーも大幅に減少した。しかし末だ原因が見付かっていないので、 今後とも動作不良が起きないよう、引き続き理学電機の責任において対処するよ う要求している。
3. 計算機及び光ケーブルによるネットワーク等
特記すべきこと無し。
Ⅱ. 可搬型プレハブハウス(TARA用プレハブ)
第3期工事として2階240m2が完成した。大部屋1、中部屋2、小部屋 12、和室1その他男子用及び女子用トイレも設けた。計算機室の脇にあった仮 眠室は2階に移す予定である。詳しくは坂部貴和子が記載した本号43頁を参照 されたい。
Ⅲ. 出資企業の増加について
今回塩野義製薬㈱が出資企業となった。参加者は(敬称略)佐藤友宏、岬真太 郎、小川政義の3氏である。
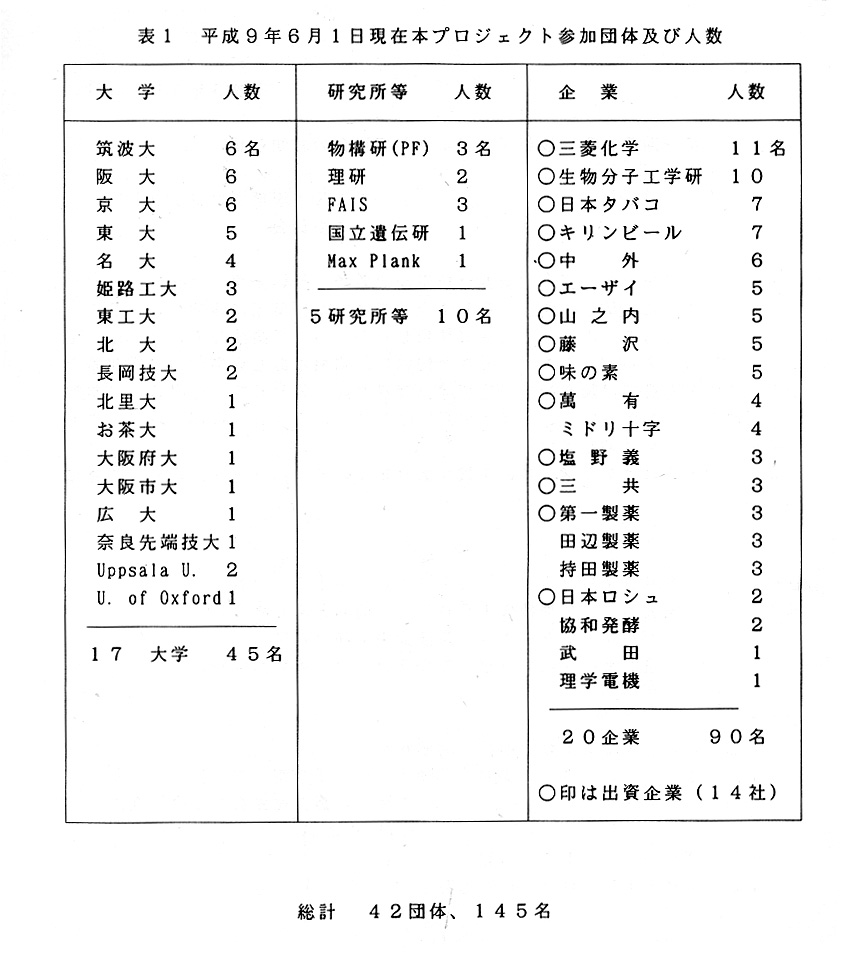
平成9年6月1日現在の本プロジェクトは;
参加企業20企業90名、その内出資企業14社、17大学45名、5研究所10名である。
総計すると42団体145名である。
各団体毎の参加人数を表1に示す。

Ⅶ. 各種委員会報告
1. 編集員会
第6回編集委員会を平成9年6月6日(金)18時よりTARAハウスにて開 催した。構造生物Vol.3,No.2の原稿の最終チェックならびに印刷等のスケジュー ルの確認が行われた。続いて次号の内容についての検討が行われ、執筆依頼者及 ぴ各委員の役割分担が決定された。
2. 行事委員会
平成9年4月28日TARAハウスにおいて「Xsightの説明トレーニング」が 行われ、20名が参加した。詳細は本号52頁の三捕圭子氏の記事を参照された 又6月26日講習会が開催されるのでそのお知らせを次頁に示す。
Ⅷ. 業績紹介
論文中にTARAのメンバー或いはTARAに謝意等を表明され、送付されて きた論文のリストを記載する。いずれ論文数が増えてきたら独立に欄を設けるベ きであるが、まだ始まったばかりで数も少ないのでここにまとめる。尚、本プロ ジェクトのメンパー名と所属を文頭に掲げた。
1. 田仲可昌(筑波大)
Group-1 introns in the cytochrome c oxidase genes of Dictyostelium
discoideum: two related ORFS in one loop of a group-J jntron, a
coxl/2hybrid gene and an unusually large cox3 gene.
Curr Genet 31:80-88, (1997).
Shinji Ogawa, Kuniko Matsuo, Kiyohiko Angata, Kaichiro Yanagisawa,
Yoshimasa Tanaka.
Summary
The DNA sequences of cytochrorme oxidase (subunits l, 2 and 3) genes of the cellular slime mold Dictyostelium discoideum mitochondria were deter -mined. The genes for subunits I and 2 have a single continuous ORF (COX1/2) which contains four group-1 introns. The insertion sites of the two group-1 introns (DdOX1/2.2 and DdOX1/2.3) coincide with those of fungal and algal group-1 introns, as well as a liverwort group-1 intron, in the cytochrorme oxidase subunit 1. Interestingly, jntron DdOXl/2.2 has two free-standing ORFS in a loop (L8) which have similar amino-acid sequences and are homologous to bi4 DNA endonuclease (1-Sce II) and bi4 RNA maturase found in group-1 introns of Saccharomyces cerevisiae mitochondrial DNA. Two group-1 introns (DdOX1/2.3 and DdOX1/ 2.4) also have a free-standing ORF in loop I and loop 2, respectively. These results show that these group-1 introns and the intronic ORFS have evolved from the same ancestral origin, but that these ORFs have been propagated independently.
Key words : Dictyostelium discoidceum , Mitochondrial DNA , Group-l intron , Cytochrome oxidase
TARAに関する表現
foot note: Y.Tanaka; Center for Tsukuba Advanced Research Alliance
(TARA researcher for Sakabe projecy), Universlty of Tsukuba, Tsukuba,
lbaraki 305, Japan.
Acknowledgments: This study was partly supported by..........and by the
Sakabe project of TARA (Tsukuba Advaced Research Alliance) of this University
2. 田仲可昌(筑波大)
Localization of a DNA Topoisomerase 11 to Mitochondria in Dictyostelium
discoideum: Deletion Mutant Analysis and Mitochondrial Targeting Signal
Presequence.
Journal of Plant Research, 110, 65-75, (1997).
Kayoko Komori 1,3, Fumlakl Maruo1 Takahlro Morio1 Hideko Urushihara1 and Yoshimasa Tanaka1,2
1Institute of Biologjcal Sciences, University of Tsukuba, Tsukuba, Ibaraki, 305 Japan.
2Center for TARA, University of Tsukuba, Tsukuba, Ibaraki, 305 Japan.
3Present address: Department of Molecular Biology, Biomolecular Engjneering Research Institute (BERI), Furuedai 6-2-3, Suita, Osaka, 565 Japan.
Summary
DNA topoisomerase 11 of Dictyostelium discoideum (TOpA), the gene (topA) encoding which we cioned, was shown to have an additional N-terminal regjon which contains a putative mitochondrial targeting signal prese- quence. We constructed overexpression mutants which expressed the wild- type or the N-terminally deleted enzyme, and examined its localization dy jmmunofluorescence microscopy and proteinase K digestlon experiment. These experiments revealed that the enzyme is located in the mito- chondria by virtue of the additional N-terminal regjon. Furthermore, in the cell extract depleted the enzyme by jmmunoprecipjtation, nuclear DNA topoisomerase 11 activity was not decreased. These results confirmed that TOPA is located in the mitochondria, even though its amino acid sequence is highly similar to those of nuclear type topoisomerase 11 of other organisms. Thus. this report is the first to establish the location of the mitochondrial targeting signal presequence in DNA topoisomerase 11 and in protelns of D. discoideum directly by analyzing deletion mutants.
Key words: Dictyostelium discoideum - DNA, topoisomerase II Localization - Mitochondria - Signal sequence
TARAに関する表現
foot note: 2: Tsukuba Advanced Research Alliance (TARA Researcher for the Sakabe project)
Acknowledgments: This work was partly supported by the Sakabe project of
TARA (Tsukuba Advaced Research Alliance) of this University.
3.神谷信夫(理研)、佐々木教祐(名大)、渡辺信久(KEK)、坂部知平(筑波大)、坂部貴和子(名大)
Time-Resolved Protein Crystal lography with Large-Angje Oscillations:
an Application of a Protein Data-Collection System Using the Weissenberg
Technique and a Large-Format Imagjng Plate.
Journal of Synchrotron Radiation, 4, 14-16, (1997).
N. Kamiya1, K. Sasaki2, N. Watanabe3, N. Sakabe4, K. Sakabe5.
1The Institute of Physical and Chemical Research (RIKEN), Hirosawa 2-1, Wako 351-01, Japan,
2 College of Medical Technology, Nagoya University, Higashi, Nagoya 461, Japan.
3Photon Factory, National Laboratory for High Energy Physics, Oho, Tsukuba 305, Japan.
4Institute of Applied Biochemistly and TARA, University of Tsukuba, Ten-no-dai, Tsukuba 305, Japan.
5Department of Chemistry, Nagoya University, Chikusa, Nagoya 464, Japan.
Summary
A diffraction-intensity data-collection system with synchrotron radiation X-rays utilizing the screenless Weissenberg technique and incorporating a large-format imaging plate is one of the most suitable apparatus for time-resolved protein crystallography with larger angle oscillations than hitherto described. The time resolution and data quality of the system have been tested using a tetragonal lysozyme crystal as a test sample in a flow-cell experiment at the bending-magnet beamline 18B at the Photon Factory, and a time resolution of 15 min is confirmed.
Keywords: Large-angle oscillations; Weissenberg technique; imaging plates; short-wavelength X-rays; Photon Factory; tetragonal lysozyme; time-resolved studies.
TARAに関する表現
footnote: Guest researcher for the Sakabe project of TARA.
4. 坂部責和子(名大)、佐々木教祐(名大)、渡辺信久(KEK)、鈴木守(KEK)、坂部知平(筑波大)
Large-Format Imaging Plate and Weissenberg Camera for Accurate Protein
Crystallographic Data Collection Using Synchrotron Radiation.
Journal of Synchrotron Radiation, 4, 136-146, (1997).
K. Sakabe1, K. Sasak2, N. Watanabe3, M. Suzuki3, Z. G. Wang3,6, J. Miyahara4,7, and N. Sakabe5.
1Department of Chemistry, Faculty of Science, Nagoya University, Nagoya 464, Japan.
2College of Medical Technology, Nagoya University, Higashi, Nagoya 461, Japan.
3Photon Factory, National Laboratory for High Energy Physics, Tsukuba 305, Japan.
4Fujj Photo Fiim Company Ltd, Kanagawa 258, Japan.
5Institute of Applied Biochemistry and Tsukuba Advanced Research Alliance (TARA), University of Tsukuba, Ibaraki 305, Japan.
6Present address: Beijjng Synchrotron Radiation Facility, Institute of High Energy Physics, Chinese Academy of Sciences, Bin 2-7, PO Box 918, Beijng 100039, People's Republic of China.
7Present Address: Fuji Photo Film Company Ltd., Nishiazabu, Minato, Tokyo 106, Japan.
Summary
Off-line and on-line protein data-collection systems using an imaging plate as a detector are described and their components reported. The off -line scanner IPR4080 was developed for a large-format imagjng plate 'BASIII' of dimensions 400X400 mm and 400X800mm. The characteristics of this scanner are a dynamic range of 105 photons pixel-1, low back-ground noise and high sensitivity. A means of reducing eiectronic noise and a method for finding the origin of the noise are discussed in detail. A dedicated screenless Weissenberg camera matching IPR4080 with synchro- tron radiation was developed and installed on beamline BL6B at the Photon Factory. This camera can attach one or two sheets of 400x800mm large-format imagjng plate inside the film cassette by evacuation. The positional reproducibility of the imaging piate on the cassette is so good that the data can be processed by batch job. Data of 93% complete- ness up to l.6A resolution were co]Iected on a single axis rotation and the value of Rmerge becomes 4% from a tetragonal lysozyme crystal using a set of two imaging-plate sheets. Comparing two types of imaging plates, the signal-to-noise ratio of the ST-VIP-type imaging plate is 25% better than that of the BASIII-type imaging plate for protein data collection using 1.0 and 0.7Å X-rays. A new on-line protein data-collection system with imaging plates is specially designed to use synchrotron radiation X-rays at maximum efficiency.
Keywords: imaging plates; Weissenberg cameras; macromolecular crystallography; data-collection systems.
TARAに関する表現
footnote: Guest researcher for the TARA Sakabe project
5. 水野 洋(農業生物資源研、筑波大)
Crystallization and Preliminary X-Ray Crystallographic Study
of Streptomyces olivaceoviridis E -86 β-Xylanase
J. Biochem., 121, 826-828, (1997)
Zui Fujimoto1, Hiroshi Mizuno1, Atsushi Kuno2, Shigeki Yoshida2 Hideyuki Kobayash3, and Isao Kusakabe2.
1Department of Biotechnology, National Institute of Agrobiological Resources, 2-1-2 Kannondai, Tsukuba, Ibaraki 305;
2Institute of Applied Biochemistry, University of Tsukuba, Tsukuba, lbarahi 305;
3National Food Research Institute. Tsukuba, Ibaraki 305
Summary
β-Xylanase from Streptomyces ol ivaceoviridis E-86 has been crystal- l ized by the hanging drop vapor diffusion method from 25% saturated ammonium sulfate and 2% Mcllvaine buffer, pH 5.7. The crystals diffract to at least 1.9Å resolution, and belong to space group P212121, with unit-cell dinaensions of a=79.6A, b=95.2Å, and c=140.3Å. There are probably two xylanase molecules (MW =45K) per asymmetric unit.
Key words: crystallization, Streptomyces olivaceoviridis, X-ray crystallography, β-xy-lanase.
TARAに関する表現
H.M. is a member of the TARA(Tsukuba Advanced Research AIliance)Project of the University of Tsukuba, Japan.
6.竹中章郎(東工大)
Crystallization of Eukaryotic E3, Lipoamide Dehydrogenase, from Yeast,
for Exhibiting X-Ray Diffraction beyond 2. 5A Resolution, and Preliminary
Structure Analysis*.
J. Biochem, 121, 1-4, (1997).
1Tomohiko Toyoda, Takeshi Sekiguchi2, and Akio Takenaka3.
1Department of Life Science, Faculty of Bioscience and Biotechnology, Tokyo Institute of Technoiogy, Midori-ku, Yohohama 226.
2Department of Fundamental Science, Faculty of Science and Technology, Iwaki Meisei University, Iwaki, Fukushima 970
Summary
Lipoamide dehydrogenase, which is a common component of α-keto acid dehydrogenase complexes, has been highly purified from yeast (Saccharomyces cerevisiae) to reveal its structure at higher resolution. New crystals obtained by a desalting method exhibited diffraction beyond 2.5Å resolution. The cell dimensions are a=97. 1, b=158.7, and c=67.9Å, and the space group is P212121. There is a dimeric enzyme in the asymmetric unit. The crystal structure was solved by means of the molecular-replacement technique and refined in a preliminary manner.
Key words: crystailization, Iipoamide dehydrogenase, X-ray study.
TARAに関する表現
footnote:*This work was supported in part by.......and by the Sakabe
project of TARA(Tsukuba Advaced Research Alliance), University of Tsukuba, Japan.
7. 田中信夫(東工大)
The 2. 8A structure of hydroxylamine oxidoreductase from a nitrifyjng
chemoautotrophic bacterium, Nitrosomonas europaea.
Nature Structural biology, 4, 276-284, april (1997).
Noriyuki Igarashi, Hideakl Moriyama, Taketomo Fujiwara, Yoshihiro
Fukumori and Nobuo Tanaka
Department of Life Science, Faculty of Bioscience and Biotechnology,
Tokyo Institute of Technology, 4259 Nagatsuta,Midori-ku, Yokohama 226,
Japan.
Summary
The 2.8A crystal structure of hydroxylamine oxidoreductase of a nitrifying chemoautotrophic bacterium, Nitrosomonas europaea, is described. Twenty-four haems lie in the centre bottom of the trimeric molecule, localized in four clusters within each monomer. The haem clusters within the trimer are aligned to form a ring that has inlet and outlet sites. The inlet is occupied by a novel haem, P460, and there are two possible outlet sites per monomer formed by paired haems lying within a cavity or cleft on the protein surface. The structure suggests pathways by which electron transfer may occur through the precisely arranged haems and provides a framework for the interpretation of previous and future biochemical and genetic observations.
TARAに関する表現
Acknowledgements: One of authors, N.T is a member of the TARA project of Tsukuba University, Japan.
8. 畑安雄(京大)、田中信夫(東工大)
The Crystal Structure of Zinc-Containing Ferredoxin from the Thermo-acidophilic Archaeon Sulfolobus sp. Strain 7#,
Biochemistry, Vol.36, 1505-1513, (1997).
Tomomi FujiI1, Yasuo Hata1, Masato Oozeki2, Hideaki Moriyama2, Takayoshi Wakagi2, Nobuo Tanaka2, aud Tairo Oshima2,3,
1Institute for Chemical Research, Kyoto University, Uji, Kyoto 611, Japan.
2Department of Life Science, Faculty of Bioscience and Biotechnology, Tokyo Institute of Technology, Nagatsuta, Yokohama, Kanagawa 226, Japan
3Present address: Department of Biotechnology, The University of Totyo, 1-1-1 Yayoi, Bunkyo-ku, Tokyto 113, Japan.
Summary
The crystal structure of ferredoxin from the thermoacidophiilicarchaeon Sulfolobus sp. strain 7 was determined by multiple isomorphous replace- ment supplemented with anomalous scattering effects of iron atomsin the Fe-S clusters, and refined at 2.0A resolution to a crystallographic R value of 0.173. The structural model contains a polypeptide chain of 103 amino acid residues, 2[3Fe-4S] clusters, and 31 water molecules; in this model, the cluster corresponding to cluster II in bacterial dicluster ferredoxins loses the fourth iron atom although it may originally be a [4Fe-4S] cluster. The structure of the archaeal ferredoxin consists of two parts: the core fold part (residues 37-103) and the N-terminal extellsion part (residues 1-36). The "core fold" part has an overall main-chain folding common to bacterial dicluster ferredoxins, containing two clusters as the active center,two α-helices near the clusters, and two sheets of two-stranded antiparallel β-sheet (the terminal and central β-sheets). The "N-terminal extension" part is mainly formed by a one-turn α-helix and a three-stranded antiparallel β-sheet. The β-sheet in the N-terminal extension is hydrogen-bonded with the terminal β-sheet in the core fold to form a larger β-sheet. The distinct structural feature of this archaeal ferredoxin lies in the zinc-binding center where the zinc ion is tetrahedrally ligated by four amino acid residues (His 16, His 19, and His 34 from the N-terminal extension, and Asp 76 from the core fold). The zinc ion in the zinc- binding center is located at the interface between the core fold and the N-terminal extension, and connects the β-sheet in the N-terminal extension and the central β-sheet in the core fold through the zinc ligation. Thus, the zinc ion plays an important role in stabilizing the structure of the present archaeal ferredoxin by connecting the N-terminal extension and the core fold, which may be common to thermoacidophilic archaeal ferredoxins.
TARAに関する表現
footnote:* This research was supported in part by..........and by the Sakabe project of TARA(Tsukuba Advaced Research Alliance), University of Tsukuba, Japan.
9. 畑安雄(京大)
The Refined Crystal Structure of Bacillus cereus Oligo-1, 6-glucosidase
at 2.0 A Resolution: Structural Characterization of Proline-substitution
Sites for Protein Thermostabilization.
J. Mol. Biol., 269, 142-153,(1997).
Kunihiko Watanabe1, Yasuo Hata2, Hidekazu Kizaki1, Yukiteru Katsube3 and Yuzuru Suzuki1.
1Department of Agticultural Chemistry, Kyoto Prefectural Universlty, Shimogamo, Sakyo, Kyoto 606, Japan.
2Institute for Chemical Research, Kyoto University, Uji, Kyoto 611, Japan
3Institute for Protein Research, Osaka University, Suita, Osaka 565, Japan.
Summary
The crystal structure of oligo-1, 6-glucosidase (dextrin 6-a -glucano- hydroiase, EC 3. 2.1.10) from Bacillus cereus ATCC7064 has been refined to 2.0Å resolution with an R-factor of 19.6% for 43,328 reflections. The final model contains 4646 protein atoms and 221 ordered water molecules with respective root-mean-square deviations of 0.015 A for bond lengths and of 3.166° for bond angles from the ideal values. The structure consists of three domains: the N-terminal domain (residues I to 104 and 175 to 480), the subdomain (residues 105 to 174) and the C-terminal domain (residues 481 to 558). The N-terminal domain takes a (β/α)s-barrel structure with additions of an α-helix (Nα6') between the sixth strand Nβ6 and the sixth helix Na6, an α-helix (Nα7') between the seventh strand Nβ7 and the seventh helix Nα7 and three α-helices (Nα8' Nα8" and Nα8"') between the eighth strand Nβ8 and the eighth helix Nα8. The subdomain is composed of an α-helix, a three-stranded antiparallel β-sheet, and long intervening loops. The C-terminal domain has β-barrel structure of eight antiparallel β-strands folded in double Greek key motifs, which is distorted in the sixth strand Cβ6. Three catalytic residues, Aspl99, Glu255 and Asp329, are located at the bottom of a deep cleft formed by the subdornain and a cluster of the two additional α-helices Nα8' and Nα8" in the (β/α)8-barrel. The refined structure reveals the locations of 21 proline-substitution sites that are expected to be critical to protein thermostabilization from a sequence comparison among three Bacillus oligo-1, 6-glucosidases with different thermostability. These sites lie in loops, β-turns and α-helices, in order of frequency, except for Cys515 in the fourth β- strand Cβ4 of the C-terminal domain. The residues in β-turns (Lysl21, Glu208, Pr0257, Glu290, Pr0443, Lys457 and Glu487) are all found at their second positions, and those in α-helices (Asnl09, Glu175, Thr261 and Ile403) are present at their N1 positions of the first helical turns. Those residues in both secondary structures adopt φ and φ va]ues favorable for proline substitution. Residues preceding the 21 sites are mostly conserved upon proline occurrence at these 21 sites in more thermostable Bacillus olig0-1, 6-gluco-sidases. These structural features with respect to the 21 sites indicate that the sites in β- turns and α-helices have more essential prerequisites for proline substitution to thermostabilize the ptotein than those in loops. This well supports the previous finding that the replacement at the appropriate positions in β-turns or α-helices is the most effective for protein thermostabilization by proline substitution.
Keywords: oligo-1, 6-glucosidase; X-ray structure; proline residue; proline substitution; thermostability.
TARAに関する表現
Acknowledgements: Y.H. is a member of the TARA (Tsukuba Advanced
Research Ailiance) project of Tsukuba University, Japan.
10. 三木邦夫(京大)
Crystallization and preliminary X-ray charachterization of chaperonin-
60 from paracoccus denitrificans.
J. Crystal Growth 168, 297-300, (1996).
Takaaki A Fukami1, Yoshio Takasuga2, Masato Sumi2, Masafumi Yohda3, Masasuke Yoshida2, Kunio Miki1,
1Department of Chemistry, Graduate School of Science, Kyoto University, Sakyo-ku, Kyoto 606-01, Japan.
2Research Laboratory of Resources Utilization, Tokyo Institute of Techn ology, Nagatsuda, Midori-ku, Yokohama 226, Japan.
3The Institute of Physical and Chemical Research, Wako, Saitama 351-01, Japan.
Summary
Chaperonin-60(cpn60) from Paracoccus denitrificans has been crystallized using polyethyleneglycol(PEG) 6000 and LiCl as precipitants. The crystals diffract up to 3.0Å resolution by using synchrotron radiation. They belong to the tetragonal space group P42212 with unit cell parameters of a=b=284Å, c=152Å. Seven subunit molecules of cpn60 seem to exist in the asymmetric unit of the crystal. A complete diffraction data set was collected using a Weissenberg camera system attached to the synchrotron radiation source and merged up to 3. 2Å resolution. Self- rotation functions calculated show the existence of a local 7-fold axis, suggesting the 7-fold symmetry ring assembly.
TARAに関する表現
Acknowledgements : --- The work on synchrotron radiation was also partly
supported by the Sakabe project at the TARA (Tsukuba Advanced Research
Alliance) center, University of Tsukuba.